| Welcome to dbPSP |
 dbPSP version 2.0 - 06/25/2019 dbPSP version 2.0 - 06/25/2019 Features Features
 dbPSP version 1.0 - 04/04/2015 dbPSP version 1.0 - 04/04/2015 Features Features
|
|
|
As one of the most ubiquitous and important protein post-translational modifications (PTMs) with tremendous studies, phosphorylation regulates a wide variety of biological processes, such as cell cycle and signal transduction (Cohen, 1982; Ptacek and Snyder, 2006; Jin, et al., 2012). Although eukaryotic protein phosphorylation has been extensively studied, only limited information is available for protein phosphorylation in prokaryotes. In contrast with eukaryotic phosphorylation that mainly occurs on serine (S), threonine (T), and tyrosine (Y), prokaryotic phosphorylation also occurs on several other types of amino acids, including arginine (R), histidine (H), cysteine (C) and aspartic acid (D) residues. dbPSP 2.0 (database of Phosphorylation Sites in Prokaryotes) is an updated resource for annotating protein phosphorylation sites (p-sites) in prokaryotes (bacteria and archaea). It contains 19,296 experimentally identified p-sites in 8,586 proteins from 200 prokaryotic organisms. In particular, detailed annotations for all the proteins were integrated from additional 88 public resources that cover 9 aspects as follows: (i) Taxonomy annotation; (ii) Genome annotation; (iii) Function annotation; (iv) Transcriptional regulation; (v) Sequence and structure information; (vi) Family and domain annotation; (vii) Interaction; (viii) Orthologous information and (ix) Biological pathway. We anticipate dbPSP can serve as a useful resource to study the mechanisms of phosphorylation in prokaryotes. dbPSP will be continuously maintained and updated, all data sets and annotations are freely accessed for all users. All data sets in dbPSP are made available under a Creative Commons CC 3.0 BY license (https://creativecommons.org/licenses/by/3.0/cn/). |
|
 Statistics
Statistics
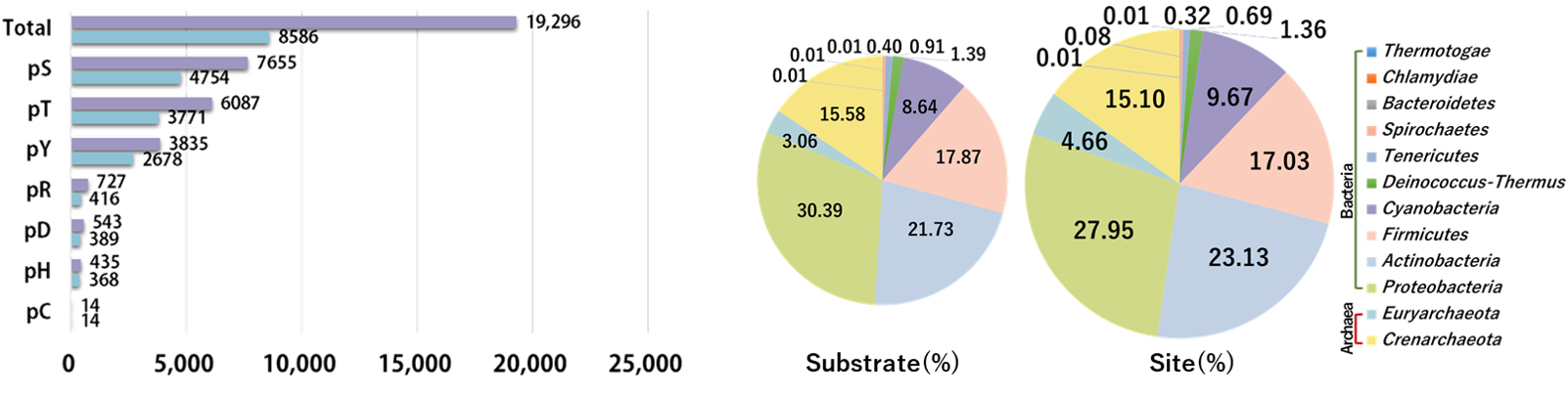 |
 Citing dbPSP
Citing dbPSP
Submitted., 2019.
Pan et al. dbPSP: a curated database for protein phosphorylation sites in prokaryotes.
Database (Oxford), 2015:bav031.





 Substrate Search
Substrate Search 